Grouping estimates by user-defined areal units
Want to compute estimates within your own areal units (spatial polygons)? All rFIA estimator functions make this task fast and easy. Simply hand your spatial polygons to the polys argument of an estimator function, like tpa or biomass, and estimates will be grouped within those spatial zones. No need to worry about projections, rFIA functions will reproject FIA data to match that of your input polygon.
## Most recent subset
riMR <- clipFIA(fiaRI)
## Group estimates by the areal units, and return as a dataframe
tpa_polys <- tpa(riMR, polys = countiesRI)
## Same as above, but return an sf mulitpolygon object (spatially enabled)
tpa_polysSF <- tpa(riMR, polys = countiesRI, returnSpatial = TRUE)
polys object must be of class SpatialPolygons (sp package), SpatialPolygonsDataFrame (sp package), or MultiPolygon (sf package). See below for help on loading data in R.
Returning estimates at the plot-level
Want to return estimates at the plot level and retain the spatial data associated with each FIA plot? Just specify returnSpatial = TRUE and byPlot = TRUE in any rFIA estimator function, and you’ve got it!
## Spatial plots with biomass
bio_pltSF <- biomass(riMR, byPlot = TRUE, returnSpatial = TRUE)
## Plot the results using default sf method
plot(bio_pltSF)
## Warning: plotting the first 10 out of 13 attributes; use max.plot = 13 to plot
## all
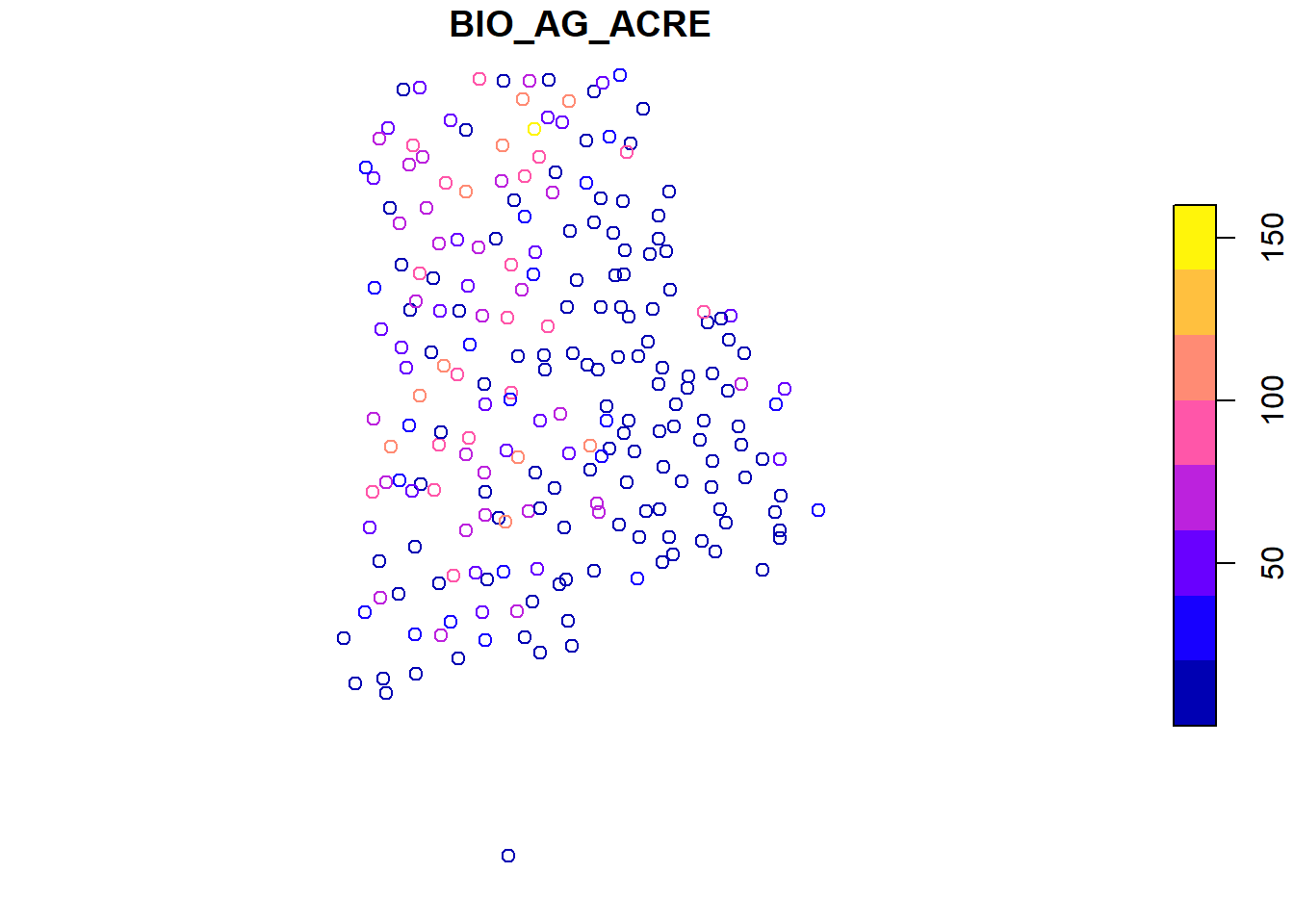
## Aboveground biomass/ acre (tons) for each plot
plot(bio_pltSF['BIO_AG_ACRE'])
Visualization
If you opted to return estimates as a spatial object (specify returnSpatial = TRUE), you can easily produce spatial choropleth maps with plotFIA:
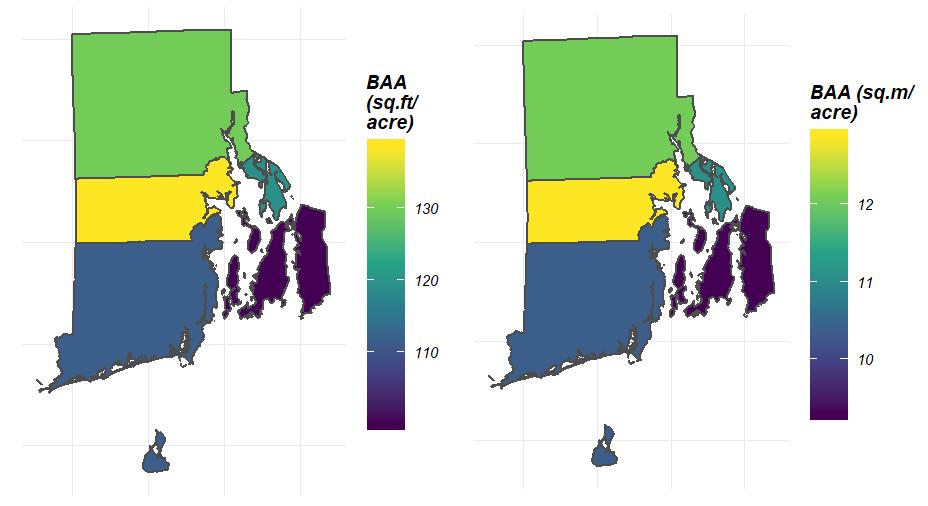
## Plot distribution of Basal area/ acre across space
plotFIA(tpa_polysSF, y = BAA, legend.title = 'BAA (sq.ft/acre)')
## You can even modify your call to y to adjust units!
## Convert sq.ft./acre to sq.m./acre
plotFIA(tpa_polysSF, y = BAA /10.7639104, legend.title = 'BAA (sq.m/acre)')
How to load spatial objects in R
There are many, many options available for working with spatial data in R. For loading multipolygon shapefiles, we recommend using readOGR from the rgdal package.
## Read a shapefile into R
## dsn: path to the directory where the shapefile components are stored
## layer: file name of the shapefile components w/ no extension
myPoly <- readOGR(dsn = 'path/to/my/folder/', layer = 'myShape')
For further reference on working with spatial data in R, we recommend these references: